Facilities
Transnational Access
Metadata & Data
Papers & Reports
Knowledge Base
Microbial Plankton
Knowledge Base
Microbial Plankton[edit]
Abstract: This deliverable is a Standard Operating Protocol (SOP) that describes methods and gathers best practice advices for sampling, enumeration and other relevant analyses of microbial communities (from viruses to ciliates) from mesocosm experiments carried out in all aquatic environments (fresh and marine waters). Quality Assurance/Quality Control (QA/QC) practices are also treated.
Use of this SOP will ensure consistency and compliance in collecting and processing microbial stock abundance and bacterial activity data from mesocosm experiments across the AQUACOSM community, in Europe and beyond.
Keywords: Microbial Analysis, Sampling, Abundance, Growth, Activity, Freshwater, Marine, Microscopy, Flow cytometry, Bacterial production, Isotopes, Enzymes, Viruses, Bacteria, Flagellates, Ciliates
Definitions and Terms[edit]
Main terms and abbreviations that will be used throughout the text
Plankton: Organisms drifting or suspended in water, consisting chiefly of minute plants or animals, but including larger forms having only weak powers of locomotion.
Biomass: The amount of living matter present in the plankton sample.
BCG: Bacterioplankton community growth. Measured by uptake of radioactively labelled substrates like the DNA base thymidine or the amino acid leucine.
TCF Thymidine conversion factor from mol thymidine assimilated to cells produced. Determined by simultaneous measurement of thymidine uptake and cell production by microscopy in seawater cultures. Alternatively, calculated from reported thymidine content in the average bacterial cell.
LCF: Leucine conversion factor. Determined as for TCF but using the amino acid leucine.
Flow cytometry: Flow cytometry is a technology used to analyse the physical and chemical characteristics of particles in a fluid as it passes through at least one laser. Cells have natural fluorescence or are often labelled with fluorescent markers so that light is first absorbed and then emitted in a band of wavelengths. The fluorescence as well as other characteristics of single particles such as their relative granularity, and its internal complexity can be quickly examined (up to thousands of cells per second) and the data gathered are processed by a computer.
Viruses: A small infective agent that typically consists of a nucleic acid molecule in a protein coat is too small to be seen by light microscopy that is able to multiply only within the living cells of a host organism. Viruses can infect all types of life forms, from animals and plants to microorganisms, including bacteria and archaea.
Prokaryotes: Usually unicellular organisms, sometimes multi cellular organisms, that lack a distinct nucleus, mitochondria, or any other membrane-bound organelle due to the absence of internal membranes. https://en.wikipedia.org/wiki/Prokaryote - cite_note-NCSU-1 The word prokaryote comes from the Greek πρό (pro) "before" and κάρυον (karyon) "nut or kernel". Bacteria are among the best-known prokaryotic organisms.
Eukaryotes: Any cell or organism that possesses a clearly defined nucleus. The eukaryotic cell has a nuclear membrane that surrounds the nucleus, in which the welldefined chromosomes (bodies containing the hereditary material) are located. Eukaryotic cells also contain organelles, including mitochondria, a Golgi apparatus, an endoplasmic reticulum, and lysosomes. Their name comes from the Greek εὖ (eu, "well" or "true") and κάρυον (karyon, "nut" or "kernel") eukaryotes may also be multi-cellular and include organisms consisting of many cell types forming different kinds of tissue. Animals, plants and fungi are the most familiar eukaryotes.
Protists: Any eukaryotic organism that is not an animal, plant or fungus. While exceptions exist, they are primarily microscopic and unicellular, or made up of a single cell. The cells of protists are highly organized with a nucleus and specialized cellular machinery called organelles.
A general description for water sampling will be covered under the Water Chemistry SOP (Version 2.0).
Materials and Reagents[edit]
Table 1-2: Reagents and recipes used in different methods
| Name and concentration | Composition | Storage |
|---|---|---|
| Acid Lugol’s Iodine |
|
Fume Cabinet/Hood |
| Formaldehyde (37% by volume) | Fume Cabinet/Hood | |
| Ethanol (96% or 99%) |
|
Solvent cabinet |
| Buffered sucrose formation (Buffered Formalin) | ||
| Glutaraldehyde (25%, for electron microscopy quality) | Fridge (4°C) | |
| SYBR Green I (other stains) |
|
Freezer (-20°C) |
| Paraformaldehyde (PFA) solution (2% final concentration in the sample) |
|
Freezer (-20°C) |
| p-phenylenediamine ant-fade mounting solution |
|
Freezer (-20°C) |
| Acridine orange |
|
Fridge (4°C) maximum 8 weeks |
| Immersion oil, low fluorescent |
|
Room temperature (4-40°C) |
| Fluorescent beads, standard |
|
Fridge (4°C) |
| TCA, Trichloracetic acid |
Health and Safety – Safe Disposal[edit]
Please see the SOPs: Water Chemistry (section 5 Water Chemistry) and Phytoplankton for general health and safety instructions regarding sampling and general lab procedures and best practices.
For specific hazardous materials and reagents mentioned in this document, such as SYBRGreen I, Glutaraldehyde, PFA, etc. please, refer to the appropriate COSHH forms by the manufacturer regarding handling and safe disposal.
Always follow the safety instructions and risk assessment protocols of the lab.
Sample Integrity – Sampling – Best Practices[edit]
Methods[edit]
Flow cytometers allow the analysis of particles of a maximum size of about 50-55 µm. For bigger cells (e.g. dynophlagellates, large diatoms and large colonies) there are valid alternative methods such as microscopy, which is described in this chapter, and flowcam analysis.
Viruses[edit]
It is recommended to use Flow Cytometry (FCM) for the counting of viruses due to the small size of viral like particles and the photobleaching of the DNA stains that are used. However, an Epifluorescence microscope can be used if a flow cytometer is not available.
i) Microscopy
Fix samples with glutaraldehyde (0.5% final concentration) for approximately 30 min at 4°C. Prepare the microscope slides within 4 h after fixation.
If it is not possible to prepare the slides within 4 h after fixation, flash freeze the samples in liquid nitrogen and store at -80°C until slide preparation.
Prepare the appropriate working solutions of SYBRGreen I and p-phenylenediamine anti-fade mounting medium.
Gently thaw the samples (if have been frozen).
Use 0.02 μm Anodisc or polycarbonate filters to filter the appropriate volume of the sample.
After filtration, dry the filters completely either by gently rubbing against a Kimi wipe or using a heating block.
Stain the filters with SYBRGreen I, dry them again and then place them on a microscopy slide. Place the appropriate amount of the p-phenylenediamine anti-fading mounting medium and put the slide cover.
To view the slide and count the viruses, use a 100x fluorescence oil-immersion objective with immersion oil. SYBR Green I binds to dsDNA and is excited with a maximum at 488 nm. A wide BP blue-excitation filter and long-pass (LP) green-emission filter will optimize observation of the cells and viruses.
A detailed protocol on viral and prokaryotic cells analysis with epifluorescence microscopy has been published by Patel et al. (2007)[1].
ii) FCM
Fix samples with glutaraldehyde (0.5% final concentration) for approximately 30 min at 4°C. Flash freeze the samples in liquid nitrogen and store at -80°C until analysis.
Before analysis, thaw samples gently and dilute to appropriate concentration for the instrument (for example, less than 700 events/sec for BD FACSCalibur) using Tris-EDTA buffer at pH 8.0.
Stain with SYBRGreen I (e.g. SYBRGreen I, ThermoFisher, S7563) to a final dilution of 5 x 10-5 of the commercial solution (e.g. use 5 μL of the 50x working dilution – mentioned at Table 1-2– at 495 μL of diluted sample). Incubate at 80°C for 10 min in the dark. Allow sample to cool down for 5 min prior to analysis.
Use a laser providing blue light (488 nm) and set trigger on green fluorescence. Group, detect and enumerate various groups of viruses based on difference in green fluorescence. Most of the times, viruses are separated in three groups based on their fluorescence.
For more information regarding the viral analysis on FCM refer to Brussaard et al. (2004)[2] and Mojika et al.(2014)[3].
Heterotrophic Bacteria[edit]
iii) Microscopy
Fix samples with 37% formaldehyde. A microscopic slide should be prepared within 7 days. Microscopic slides can be stored at -20°C for 70 days before analysis. Apply a GF/C filter on a filter support of a multifilter unit wetted with MilliQ water. Place a black 0.2 µm polycarbonate filter with shiny side up on top of the GF/C filter. Additional filter pore sizes may also be appropriate, i.e. 3.0 µm for discriminating amongst particle-associated and free-living bacteria (Hobbie et al.[4], Azam et al[5]), particularly when algal or zooplankton densities are high. Apply vacuum to make the filters stick to the support. Shake the sample for 10 s and take out the appropriate volume of sample for a proper density of cells (the volume depends on the trophic status of the environment). Wash the tip in MilliQ water between transfers. Filter the sample dry with -13 kPa vacuum. Remove vacuum. Add 15 drops of Acridine orange (AO) stain (3 mg mL1) or l−1 DAPI (4,6-diamidino-2-phenylindole) through a 0.2 µm filter (e.g. Acrodisc). Incubate for 10 min, label the glass slide in the meantime. Filter until surface is dry. Remove vacuum. Wash with 1 mL MilliQ water, filter dry. Dry filter in the air holding it with forceps for 45 s. Mount on glass slide with immersion oil spread on a spot. Add a drop of immersion oil on top of the filter and apply a cover slip. Analyse the sample in an epifluorescence microscope with filter set (FS 09, 450-490, FT 510, LP 520) and 63x PlanApochromat objective (Hobbie et al. 1977[4]). Preferably take 5 images per sample with a high resolution black and white camera. Count cell numbers, morphology and size by a neural network analysis software like LabMicrobe (BioRas, Denmark, Blackburn et al. 1998)[6]. Manual counting using ocular square grids like the Miller Square grid can also be used. The magnification factor between counted area and total filtered areas needs to be determined for each method. Fluorescent beads are used to calibrate the system regarding particles counted and their size. The cell morphologies of cocci, rods and vibroids in the size range 0.2-2 µm are counted as bacteria. Sample images of these are used to train the neural network. Cell volumes can be used to calculate bacterial carbon density and biomass (Norland 1993)[7].
iv) FCM
Fix samples with glutaraldehyde (0.5% final concentration) for ca 30 min at 4°C. Flash freeze the samples in liquid nitrogen and store at -80°C until analysis. Before analysis, thaw samples gently keeping them at 4°C on an freezer block, dilute to appropriate concentration. Stain with SYBRGreen I (e.g. MolecularProbes, Eugene, Oregon, USA) and incubate in the dark at room temperature for 10-20 min at a final concentration of 1:10000 of the stock SYBRGreen solution (Marie et al. 1999)[8]. For this, prepare a 1:100 solution of SYBRGreen diluted in 0.2 µ filtered DMSO (1µL of stock solution in 99 µL of DMSO) and add 10 µl of this solution per 1 ml of sample. Use a laser providing blue light (488 nm) and set trigger on green fluorescence. Especially for freshwater samples background noise is high due to inorganic suspended particles which also are positive in green fluorescence. Carefully set the trigger at the border of the smallest positive population and dilute as much as possible your sample. Add 0.2 µm of fluorescent beads (e.g. Thermo Fisher Scientific, Whaltam, Massachussets, USA) to compare relative sizes. Group, detect and enumerate various populations of bacteria based on difference in green fluorescence (BL1) and scatter properties (SSC).
Autotrophic bacteria and nanophytoplankton:[edit]
v) Microscopy
For the enumeration of autotrophic bacteria (Synechococcus spp.) and nanoflagellates, samples are fixed with 1.8% formaldehyde buffered with sodium tetraborate decahydrate and filtered for particle removal through 0.45 μm filters. Fixation is allowed for 1 h in the dark and at 4°C. Subsequently, samples are stained with 0.2 mg l−1 DAPI (4,6-diamidino-2-phenylindole) for 10 min and filtered through 0.6 μm black polycarbonate membranes (Porter and Feig 1980[9]). Filters are mounted on glass slides and stored at -20°C until analysis. Enumeration is performed using epifluorescence microscopy under UV-excitation. Autotrophic bacteria and nanoflagellates are distinguished by their orange and red fluorescence, respectively, observed under blue light excitation; at least 50 fields are counted for each sample. Nanoflagellates are separated into four size classes (< 2 μm, 2-5 μm, 5-10 μm and >10 μm) using an ocular micrometer. An ellipsoid shape is assumed for nanoflagellates and the biovolume is calculated separately for each size class using the measured dimensions. Nanoflagellate biomass is calculated using the conversion factor 183 fg C μm-3 described by Caron et al. (1995)[10]. Synechococcus abundance data are converted into C biomass using 250 fg C cell-1 (Kana and Glibert, 1987)[11].
vi) FCM
The best analysis of autorophic bacteria and nanophytoplancton analysis is obtained from fresh samples but, if you need to fix them before analysis, follow the same protocol as described for heterotrophic bacteria and viruses. Use a laser providing blue light (488 nm), and ideally a yellow laser (561 nm) if available, set trigger on red fluorescence channel and group, detect and enumerate various groups of phytoplankton (typically Synechococcus, Prochlorococcus, picoeukaryotes, nanoeukaryotes, cryptophytes) based on chlorophyll (red fluorescence), phycoerythrine (yellow/orange fluorescence) and phycocyanine (660 nm, for cryptophytes) autofluorescence and side-scatter (SSC) signals (e.g. Larsen et al. 2001)[12].
Heterotrophic Flagellates[edit]
vii) Microscopy
For the enumeration of heterotrophic nanoflagellates, samples are fixed with 1.8% formaldehyde buffered with sodium tetraborate decahydrate and filtered for particle removal through 0.45 μm filters. Fixation is allowed for 1 h in the dark and at 4°C. Subsequently, samples are stained with 0.2 mg l−1 DAPI (4,6-diamidino-2-phenylindole) for 10 min and filtered through 0.6 μm black polycarbonate membranes (Porter and Feig 1980[9]). Filters are mounted on glass slides and stored at -20°C until analysis. Enumeration is performed using epifluorescence microscopy under UV-excitation. Autotrophic nanoflagellates are distinguished from heterotrophic ones by the red fluorescence of their chloroplasts, observed under blue light excitation; at least 50 fields are counted for each sample. Nanoflagellates are separated into four size classes (<2 μm, 2-5 μm, 5-10 μm and >10 μm) using an ocular micrometer. An ellipsoid shape is assumed for nanoflagellates and the biovolume is calculated separately for each size class using the measured dimensions. Biomass is calculated using the conversion factor 183 fg C μm-3 described by Caron et al. (1995)[10].
viii) FCM
Fix samples with glutaraldehyde (0.5% final concentration) or paraformaldehyde (1% final concentration) for at least 2 h at 4°C. Flash freeze the samples in liquid nitrogen and store at -80°C until analysis. Thaw samples gently before analysis and stain with SYBRGreen I (e.g. MolecularProbes, Eugene, Oregon, USA) for at least 10 min in room temperature (Zubkov et al. 2007)[13]. Using a laser providing blue light (488 nm), set trigger on green fluorescence and discriminate HNF population(s) from nano-sized phytoplankton based on green vs. red fluorescence and from large bacteria on plots of side scatter vs. green fluorescence following the recommendations of Christaki et al. (2011)[14].
Micro-Plankton (Dinoflagellates, Ciliates)[edit]
For ciliate and dinoflagellate enumeration, samples (the sample volume depends on the trophic status of the environment) are preserved with borax-buffered formaldehyde (final concentration 2%) or with acid Lugol’s solution (final concentration 2%). The samples are stored at 4°C in the dark and examined within 3 month after collection. Before examination, samples are left to settle for about 24 h in Utermöhl chambers (Utermöhl 1931[15]) and are finally examined with an inverted microscope at 200×. The microscope may be equipped for transmitted light, phase-contrast and epifluorescence.
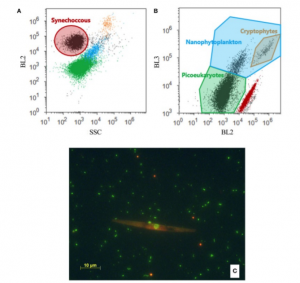
Blue light excitation (DM 500 nm dichroic mirror, BP 420 to 480 nm exciter filter, BA 515 nm barrier filter and a 100 W mercury burner) is used to detect chlorophyll autofluorescence and to distinguish plastidic from non-plastidic ciliates. A problem associated with the preservative choice is the possibility of affecting the apparent importance of loricate and aloricate ciliates since tintinnids are expected to be more robust to preservation. Buffered formaldehyde (final concentration 2%) is often used especially when we need to know about the trophic status of ciliates. Stoecker et al. (1989)[16] established that samples preserved in buffered formaldehyde lost 10 to 20% of aloricate ciliates compared to samples preserved in acid Lugol’s iodine solution, whereas Revelante & Gilmartin (1983)[17] estimated this loss to be 30 to 70%.
Processes[edit]
1) Bacterioplankton community growth (Smith and Azam 1992[18])
Wash a 50 mL polypropylene tube (e.g. Falcon™) with sample water and then collect the sample. Fill 2-4 micro vials (polypropylene, e.g. Eppendorf) with 1 ml sample water. Where many true replicates occur, 1 sample and one or fewer controls can be used. Otherwise, 3 replicate samples and one control are recommended. Wash the tip with MilliQ water and sample water between each sample type. Add 100µL ice cold 50% trichloroacetic acid (TCA) to the control samples. Vortex 3 s and invert the tube to mix.
Incubate 5 min at 2°C. Take out the total required amount of 3H-Thymidine (80 000 Ci mol-1, concentration of 12.5 µM, 1mCi mL-1) to a micro vial. Transfer 2 µL of the isotope (20 nmol dm-3 final conc.) to the wall above the liquid to each vial starting with the samples and finish with the controls. Mix all vials 3 s and note the time for incubation start. Incubate in a thermos with the in-situ temperature for 1 hour. Stop the incubation by putting the vials into a micro vial cooling-block (2°C, e.g. Eppendorf Thermostat C for 5 min.). Note the stopping time. Add 100 µL of ice cold 50% TCA to the sample tubes only. Incubate for 5 min. Samples can be stored at this temperature up to 7 days. Centrifuge the samples with necks facing outwards at 16000 × g (13000rpm) for 10 min. Remove the supernatant and any moisture in the lid or walls by a tap water vacuum evaporator with a drawn Pasteur glass pipette. Add 1ml of 5% TCA, turn the vial upside down removing any air bubble and then vortex 5 s. Centrifuge again as above. Remove the supernatant as above. Finally add 1 mL Scintillation liquid (e.g. Optiphase HiSafe) to each vial. Place in scintillation vials (5 mL size). Samples can be stored at room temperature in the dark for at least 1 month. Count samples in the tritium channel in a calibrated scintillation counter with applied quench correction. Calculate the uptake of thymidine according to the reference above. Convert to cell production by using empirical or theoretical thymidine conversion factor (Wikner and Hagström 1999[19]). Calculation to biomass production can be done by multiplying with the per cell carbon content from bacterial biomass estimates. When bacterial biomass production is wanted 3H-leucine (Perkin-Elmer Life Science Products; 170 mCi mmol-1) may be used instead of thymidine, applying the theoretical conversion factor for leucine (del Giorgio et al. 2011).
References[edit]
- ↑ Patel A., Noble R.T., Steele J.A., Schwalbach M.S., Hewson I., Fuhrman J.A. (2007) Virus and prokaryote enumeration from planktonic aquatic environments by epifluorescence microscopy with SYBR Green I. Nat. Protoc. 2: c269-276.
- ↑ Brussaard C.P.D. (2004) Optimization of Procedures for Counting Viruses by Flow Cytometry. Appl. Env. Microbl. 70: 1506-1513.
- ↑ Mojika K.D.A., Evans C., Brussaard C.P.D. (2014) Flow cytometric enumeration of marine viral populations at low abundances. Aquat. Microb. Ecol. 71: 203-209.
- ↑ 4.0 4.1 Hobbie J.E., Daley R.J. and S. Jasper (1977) Use of nucleporefilters for counting bacteria by fluorescence microscopy. Appl. Environ. Microbiol. 33: 1225-1228.
- ↑ Azam, F., Holm-Hansen, O. Use of tritiated substrates in the study of heterotrophy in seawater. Mar. Biol. 23, 191–196 (1973). https://doi.org/10.1007/BF00389484
- ↑ Blackburn N. et al. (1998) Rapid Determination of Bacterial Abundance, Biovolume, Morphology, and Growth by neural Network-Based Image Analysis. Appl. Environ. Microbiol. 64(9): 3246-3255.
- ↑ Norland S. (1993) The relationship between biomass and volume of bacteria. In Kemp P.F., B.F. Sherr, E.B. Sherr and J.J. Cole [eds.], Handbook of methods in aquatic microbial ecology. Lewis Publishers: p. 303-307
- ↑ Marie D., Partensky F., Jacquet S. and Vaulot D. (1997). Enumeration and cell cycle analysis of natural populations of marine picoplankton by flow cytometry using the nucleic acid stain SYBR Green I. Appl. Environ. Microbiol. 63: 186-93
- ↑ 9.0 9.1 Porter K.G., Feig Y.S. (1980) The use of DAPI for identifying and counting aquatic microflora. Limnol. Oceanogr. 25: 943-948. https://doi.org/10.4319/lo.1980.25.5.0943.
- ↑ 10.0 10.1 Caron D.A., Dam H.G., Kremer P., Lessard E.J., Madin L.P., Malone T.C., Napp J.M., Peele E.R., Roman M.R., Youngbluth M.J. (1995) The contribution of microorganisms to particulate carbon and nitrogen in surface waters of the Sargasso Sea near Bermuda. Deep-Sea Res. Part I 42: 943-972. https://doi.org/10.1016/0967-0637(95)00027-4
- ↑ Kana T., Glibert P.M. (1987) Effect of irradiances up to 2000mE m-2 s-1 on marine Synechococcus WH 7803-I. Growth, pigmentation and cell composition. Deep Sea Research 34: 479-516.
- ↑ Larsen A., Castberg T., Sandaa R.A., Brussaard C.P.D. Egge J., Heldal M. et al. (2001). Population dynamics and diversity of phytoplankton, bacteria and viruses in a seawater enclosure. Mar. Ecol. Prog. Ser. 221: 47-57. doi:10.3354/meps221047
- ↑ Zubkov M.V., Burkill P.H. and Topping J.N.(2007). Flow cytometric enumeration of DNA-stained oceanic planktonic protists. J. Plankton Res. 29: 79–86. doi:10.1093/plankt/fbl059.
- ↑ Christaki U., Courties C., Massana R., Catala P., Lebaron P., Gasol J.M. et al. (2011).Optimized routine flow cytometric enumeration of heterotrophic flagellates using SYBR Green I. Limnol. Oceanogr. Methods 9:329-339. doi: 10.4319/lom.2011.9.329.
- ↑ Utermohl von H. (1931) Neue Wege in der quantitativen Erfassung des Planktons. (Mit besondere Beriicksichtigung des Ultraplanktons). Verhandlungen der Int. Vereinigung fur Theor. und Angew. Limnol. 5: 567-595
- ↑ Stoecker D.K., Taniguchi A. and Michaels A.E. (1989) Abundance of autotrophic, mixotrophic, and heterotrophic planktonic ciliates in shelf and slope waters. Mar Ecol Prog Ser 50: 241-254
- ↑ Revelante N. and Gilmartin M. (1983) Microzooplankton distribution in the Northern Adriatic Sea with emphasis on the relative abundance of ciliated protozoans. Oceanol Acta 6: 407-415.
- ↑ Smith D.C. and Azam F. (1992) A simple, economical method for measuring bacterial protein synthesis rates in seawater using 3H-leucine. Mar. Microb. Foodwebs 6:107-114.
- ↑ Wikner J. and Hagström Å. (1999) Bacterioplankton intraannual variability at various allochthonous loading: Importance of hydrography and competition. Aquat. Microb. Ecol. 20: 245-260.